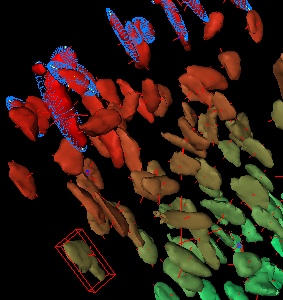
 This plugin performs connected-components labelling. It started life as an adaptation of the 3D Object Counter, for identifying particles as part of Purify. Once it became obvious that the heavily recursive algorithm was very inefficient, especially on large stacks, particle labelling was rewritten to be multithreaded and much less recursive. A further, single-threaded, non-recursive (runs in O(n) time) algorithm is available but is very heavy on RAM and only significantly outperforms the multithreaded algorithm on very large stacks, but can be faster on small stacks in some cases.
This plugin performs connected-components labelling. It started life as an adaptation of the 3D Object Counter, for identifying particles as part of Purify. Once it became obvious that the heavily recursive algorithm was very inefficient, especially on large stacks, particle labelling was rewritten to be multithreaded and much less recursive. A further, single-threaded, non-recursive (runs in O(n) time) algorithm is available but is very heavy on RAM and only significantly outperforms the multithreaded algorithm on very large stacks, but can be faster on small stacks in some cases.
Since BoneJ v1.3.10, Particle Analyser joins particles using a 3-pass non-recursive approach that maps neighbourhoods as in a graph and merges them very quickly and memory-efficiently. This fast labelling feature is now the default method. It is listed as 'Mapped' in the algorithm selection drop-down.
Major speed improvements (50×) have been made to this plugin, available only in
BoneJ2. For more details, see the
preprint describing the new approach.
A member of our group wished to study osteocyte lacunae in synchrotron microCT images, so the Particle Analyser was born from Purify's particle labelling algorithm. Other groups use Particle Analyser for soil analysis; it could be used for any porous media. Particle Analyser labels unique (unconnected) particles, then analyses each particle separately. Substantial use is made of other plugins in BoneJ, including Connectivity to get the Euler characteristic, Isosurface to get the surface area and Thickness to get the local thickness of individual particles.
Installs in the ImageJ menus under Plugins->Analyze->Particle Analyser.
- Input
- Options
- Exclude on sides: don't analyse particles touching the stack sides
- Feret Diameter: measure particles' Feret diameter (slow, brute force method).
- Moments of inertia: measure each particle's moments of inertia
- Thickness: Measure each particle's local thickness. Note: does not handle anisotropic voxels due to the underlying implementation of Local Thickness.
- Surface Area: measure particles' surface area from surface mesh
- Enclosed volume: measure volume enclosed by each particle's surface mesh
- Euler characteristic: measure each particle's Euler characteristic and connectivity
- Ellipsoids: find the best-fit ellipsoid to the particle's surface mesh
- Record unit vectors: log the unit vectors of the inertia tensor and best-fit ellipsoid's axes to the Results table
- Min Volume: smallest particle to measure or display
- Max Volume: largest particle to measure or display
- Surface resampling: how much to resample the stack while creating the surface mesh by marching cubes. The effect of this option can be visualised with Isosurface.
- Show particle stack: display an image containing labelled particles
- Show thickness stack: display the result of local thickness in a new stack
- Show centroids (3D): display particles' centroids in a 3D Viewer window
- Show ellipsoids (3D): display best-fit ellipsoids in a 3D Viewer window
- Show size stack: display an image with particles labelled by their volume
- Show surfaces (3D): display particles' surface meshes in a 3D Viewer window
- Show axes (3D): display principal axes (from moments of inertia) in a 3D Viewer window
- Show stack (3D): display the original binary stack in a 3D Viewer window
- Surface colours:
- Gradient: Each particle gets a unique colour between red and green
- Split: Each particle is coloured depending on whether it is over (red) or under (yellow) the Split value
- Split value: Particles with volume greater than this value are coloured red, otherwise coloured yellow.
- Volume resampling: amount of voxel resampling to apply when displaying the original stack
- Labelling algorithm
- Mapped: Non-recursive and memory-efficient, fastest method but with slightly higher memory requirement than Multithreaded.
- Multithreaded: use multiple cores and job chunking to reduce recursion. Fast on small stacks and if you have many CPU cores.
- Linear: Non-recursive but heavy on RAM and single-threaded. Fast on big stacks, but only if you have the memory.
- Slices per chunk: number of image slices per thread when using the Multithreaded particle-labelling algorithm. For large images (512 x 512 x 512) and above, use 2, for smaller images, 3 or 4 may be faster on your system.
- Output
- Results:
- Label: Image file name
- ID: unique particle identifier; this number is the label used for the particle in all calculations and output
- Vol: particle volume
- x Cent: x-coordinate of particle centroid
- y Cent: y-coordinate of particle centroid
- z Cent: z-coordinate of particle centroid
- SA: surface area (0 if too small for mesh to be produced; see warning log)
- Encl. Vol: Volume enclosed by surface mesh (0 if too small for mesh to be produced; see warning log)
- I1: moment of inertia around shortest principal axis
- I2: moment of inertia around middle principal axis
- I3: moment of inertia around longest principal axis
- vX: x component of unit vector of longest principal axis (a measure of orientation)
- vY: y component of unit vector of longest principal axis
- vZ: z component of unit vector of longest principal axis
- vX1 - vZ2: xyz components of the unit vectors of the middle and short principal axes
- Euler (χ): Euler characteristic of the particle
- Holes (β1): number of topological holes (handles) in the particle
- Cavities (β2): number of enclosed cavities in the particle
- Thickness: mean local thickness of particle
- SD Thickness: standard deviation of the mean local thickness of particle
- Max Thickness: maximum local thickness of particle
- Major radius: length of best-fit ellipsoid's long radius
- Int. radius: length of best-fit ellipsoid's intermediate radius
- Minor radius: length of best-fit ellipsoid's short radius
- V00 - V22: elements of the 3Ã3 rotation matrix of the best-fit ellipsoid
- Image Stacks
- Thickness image showing local thickness at each point in the input image
- Particle image showing the particle ID at each point in the input image (individual particles can be selected by thresholding for that ID value)
- Size image showing particles labelled by their volume; particles can be selected based on volume by thresholding this image
- 3D display
- If any of the 3D display options are selected, an ImageJ 3D Viewer window is opened. All features are added individually, so that by hovering the mouse over a particle's surface, you can see its ID in the ImageJ status area. Results pertaining to that particle can then be looked up in the Results table. Particles can be manipulated individually, including transforming and adjusting colour or transparency.

 This file last modified 1345hrs 29 May 2020 © Michael Doube 2004-2026 :: Designed to be interoperable and standards-compliant.
This file last modified 1345hrs 29 May 2020 © Michael Doube 2004-2026 :: Designed to be interoperable and standards-compliant.
 This plugin performs connected-components labelling. It started life as an adaptation of the 3D Object Counter, for identifying particles as part of Purify. Once it became obvious that the heavily recursive algorithm was very inefficient, especially on large stacks, particle labelling was rewritten to be multithreaded and much less recursive. A further, single-threaded, non-recursive (runs in O(n) time) algorithm is available but is very heavy on RAM and only significantly outperforms the multithreaded algorithm on very large stacks, but can be faster on small stacks in some cases.
This plugin performs connected-components labelling. It started life as an adaptation of the 3D Object Counter, for identifying particles as part of Purify. Once it became obvious that the heavily recursive algorithm was very inefficient, especially on large stacks, particle labelling was rewritten to be multithreaded and much less recursive. A further, single-threaded, non-recursive (runs in O(n) time) algorithm is available but is very heavy on RAM and only significantly outperforms the multithreaded algorithm on very large stacks, but can be faster on small stacks in some cases.
